Abstract
Introduction: Perturbations in alternative splicing have been associated with multiple pathogenic processes in cancer. We performed a genome-wide screen for differential splicing between subtypes of diffuse large B-cell lymphoma (DLBCL), demonstrate that alternatively spliced isoforms of BCAP (PIK3AP1) - an adaptor protein linking B-cell receptor (BCR) signaling to PI3 kinase activation - differ in function, and propose a mechanism by which BCAP splicing contributes to oncogenic signaling.
Methods: Samples with high read depth (>60M mapped paired-ends) RNA sequencing (RNAseq) from a prior study of DLBCL (Schmitz et al. NEJM 2018) were assayed for differential splicing with a custom bioinformatics pipeline (N = 355). Splice junction reads were quantified using MAJIQ, p-values were calculated using the rMATS statistical framework, and false discovery rate (FDR) calculated using the Benjamini-Hochberg method. Previously published RNAseq from tonsillar biopsies and IgM-stimulated peripheral B-cells (Schmitz et al. Nature 2012) was analyzed for BCAP gene expression and splicing. Knockdown-rescue experiments were performed in four activated B-cell (ABC) DLBCL cell lines (TMD8, HBL1, OCI-Ly10, Riva) and one germinal center B-cell (GCB) DLBCL cell line (WSU-WFSCCL) expressing Cas9. Genomic BCAP was knocked down using a vector co-expressing GFP and an sgRNA recognizing an exon-intron junction, and cell lines were rescued with a vector co-expressing Lyt2 (mouse CD8) and either the long (BCAP-L) or short (BCAP-S) gene isoform. For proximity labeling, we expressed a fusion protein of each isoform of BCAP linked to BioID2, a promiscuous biotin ligase, at either its N- or C-terminal. Biotinylated proteins in BCAP-BioID2-expressing Riva cells were purified and compared to proteins from controls by SILAC (stable isotope labeling by amino acids in culture)-based quantitative mass spectrometry.
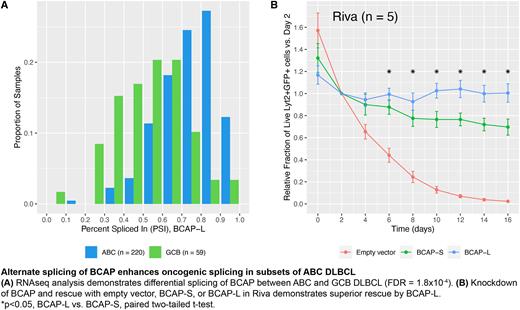
Results: Bioinformatic analysis identified BCAP as differentially spliced between gene expression (ABC vs. GCB, FDR = 1.8 x 10-4; Fig. A) and genetic subtypes of DLBCL (non-EZB vs. EZB, FDR = 0.048). No mutations were identified within the involved splice sites. Alternate transcription start sites in BCAP result in expression of either BCAP-L or BCAP-S, which differ by 178 amino acids containing a TIR domain present in BCAP-L. As a fraction of total gene expression, GCB and EZB subtypes express lower levels of BCAP-L (median 0.57 and 0.59, respectively) than ABC and non-EZB subtypes (median 0.76 and 0.74, respectively).
In normal B-cells, tonsillar germinal center cells express a proportion of BCAP-L similar to GCB DLBCL (median BCAP-L fraction = 0.56). In contrast, in vitro stimulation of peripheral B-cells led to transient upregulation predominantly of BCAP-L (maximum BCAP-L fraction = 0.87), resembling ABC DLBCL. These results suggest that increased BCAP-L fraction observed in ABC DLBCL is a transcriptional response to chronic active BCR signaling.
To test for a functional difference between BCAP isoforms, we performed knockdown-rescue experiments in five Cas9-transduced DLBCL cell lines with an sgRNA targeting genomic BCAP and rescuing with either BCAP-L or BCAP-S. Consistent with pooled sgRNA library screens, BCAP knockdown resulted in decreased viability of all tested ABC cell lines. Additionally, in Riva and OCI-Ly10 cells, BCAP-L rescue was superior to that of BCAP-S, confirming our hypothesis (Fig. B).
To further characterize differences between BCAP-L and BCAP-S, we identified protein-protein interactions of each isoform using BioID2 proximity labeling in Riva cells. This confirmed known interactions of BCAP including NCK, BTK, PLCG2, and PIK3R1 (p85). However, compared to BCAP-S, BCAP-L demonstrated increased association with components of BCR signaling including immunoglobulin kappa constant region (IGKC), CD2AP, SH3KBP1, and BLNK.
Conclusions: Our findings suggest that chronic active B-cell receptor signaling leads to preferential splicing for BCAP-L, which in turn enhances oncogenic signaling in DLBCL. These experiments also support prior hypotheses that BCAP-L is the predominant isoform of BCAP transducing external cell stimuli through localization of PI3K kinases to the plasma membrane. Further characterization of alternative splicing may potentially inform pathway dependencies and therapy in DLBCL and other malignancies.
Disclosures
Itsara:Adaptive Biotechnologies: Other: Spouse: current employee and holder of stock options. Oellerich:Merck KGaA: Consultancy, Research Funding; F. Hoffmann-La Roche Ltd: Consultancy, Honoraria; Gilead Sciences, Inc.: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal